WEHI SODA Hub
The Spatial Omics Data Analytics Hub (SODA Hub) supports WEHI's research at the cutting edge of spatial omics through the development of novel analysis tools, workflows and data management solutions.
View the Project on GitHub WEHI-SODA-Hub/wehi-soda-hub.github.io
Contact us:WEHI SODA Hub
The Spatial Omics Data Analytics (SODA) Hub was established to support and accelerate WEHI’s research using cutting-edge spatial omics technology by enabling robust data management and state-of-the-art analyses. Researchers in this field face unique challenges with large and complex multimodal datasets, and a growing ecosystem with rapidly changing technologies. The SODA Hub comprises an inter-disciplinary team with experience in research software engineering, bioinformatics, and bioimage analysis. The team is developing innovative data management, storage and analysis solutions, enabling scientists to work more efficiently with spatial omics data across a range of technologies.
The Team

The SODA Hub project is being led by joint lead investigators Professor Kelly Rogers, Professor Marie-Liesse Labat and Professor Tony Papenfuss. The technical team consists of:
- Dr Marek Cmero – Senior Research Officer (@mcmero)
- Mr Michael Milton – Research Computing Engineer (@multimeric)
- Dr Pradeep Rajasekhar – Senior Research Officer (@pr4deepr)
- Dr Lachlan Whitehead – Senior Research Officer (@DrLachie)
- Dr Julie Iskander – Head, Research Computing Platform (@jIskCoder)
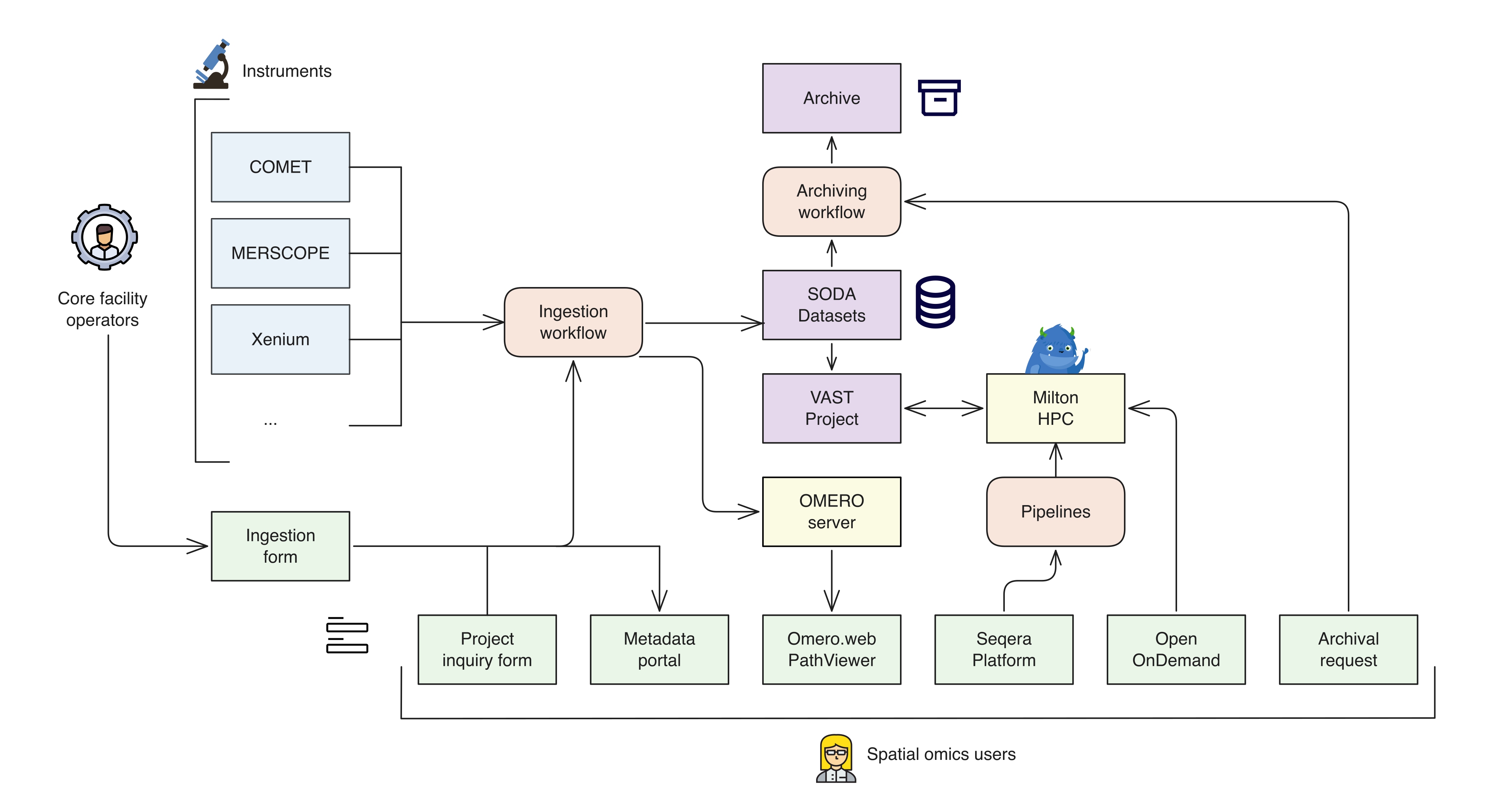
Data flows
The SODA Hub is designing a streamlined environment from data ingestion to analysis. The figure below shows how data flows through the environment, with data transfer, metadata capture, and preprocessing handled in a streamlined workflow. Spatial omics users are then able to access and analyse their spatial data in numerous ways:
- A metadata portal for validating and searching for metadata.
- OMERO.web and PathViewer for image management, visualisation and annotation.
- Seqera Platform for launching Nextflow pipelines for segmentation, phenotyping and other workflows on the local HPC.
- Virtual Desktops via Open OnDemand for working interactively with data using QuPath or other tools.
Software
SODA is building and contributing to open-source software. Please see the WEHI-SODA-Hub GitHub page for a complete list of our public repositories.
Pipelines
- spatialvpt: Segmentation and QC workflow for MERSCOPE using the vizgen-postprocessing tool via a parallelised and streamlined workflow.
- sp_segment: Cell segmentation pipeline for spatial proteomics (COMET & MIBI). Can run patched segmentation using Cellpose via SOPA. Resolves whole-cells and nuclei and calculates cell measurements.
- sp_celltype_preprocess: Prepare spatial proteomics dataset for training or prediction of cell types.
- sp_celltype_train: Train an XGBoost model on a pre-processed labelled spatial proteomics dataset.
- sp_celltype_apply: Apply a trained XGBoost model on pre-processed spatial proteomics data to predict cell types.
Analysis tools
- MibiSegmentation: A CLI for Mesmer segmentation of COMET and MIBI TIFFs used in the sp_segment pipeline. Automatically combines membrane channels.
- cellmeasurement: Groovy app for calculating cell measurements from whole-cell and nuclear masks used in the sp_segment pipeline. Performs matching of whole-cells and nuclei and automates calculation of measurements using the QuPath API.
- spatialVis: R package for downstream plots and spatial analyses for spatial proteomics. Provides functions for plotting cell segmentation and phenotyping QC metrics, as well as a report template for generating reports. Used to generate segmentation reports for the sp_segment pipeline.
Metadata tools
- rdfpytest: Framework for running SHACL tests over RDF data in Python.
- RdfNav: Utilities for navigating an RDF graph in Python.
- RdfCrate: Library for building RO-Crates using rdflib.
- OmeroCrate: Tools for uploading an RO-Crate dataset to Omero.
Funding
The WEHI SODA Hub is made possible due to generous support of The Kinghorn Foundation.